|
Presenting Author Information |
|
|
Name |
Ravi Shankar |
|
Institution |
Stanford UUniversity |
|
PI |
Mark A. Musen |
|
|
|
|
Phone Number |
4086912271 |
|
Additional Author Information |
|
|
Names and affiliations of additional authors (one per line) Marcos Martinez-Romero, Stanford University Martin J. O'Connor, Stanford University John Graybeal, Stanford University Purvesh Khatri, Stanford University Mark A. Musen, Stanford University |
|
|
Is there an additional contact person? |
No |
|
Additional information |
|
|
Please choose the topic that best fits your abstract (posters will be grouped according to your selection). Detailed session descriptions can be found in the Abstract Guidelines. |
Software, Analysis, & Methods Development |
|
Please consider my abstract for a (See Presentation Guidelines) |
Either poster or presentation |
|
Abstract Information |
|
|
Poster presentations may be submitted electronically in order to reach a wider audience and be available after the All hands meeting. Do you plan to submit your poster as a digital submission in addition to bringing a physical copy? |
Yes |
|
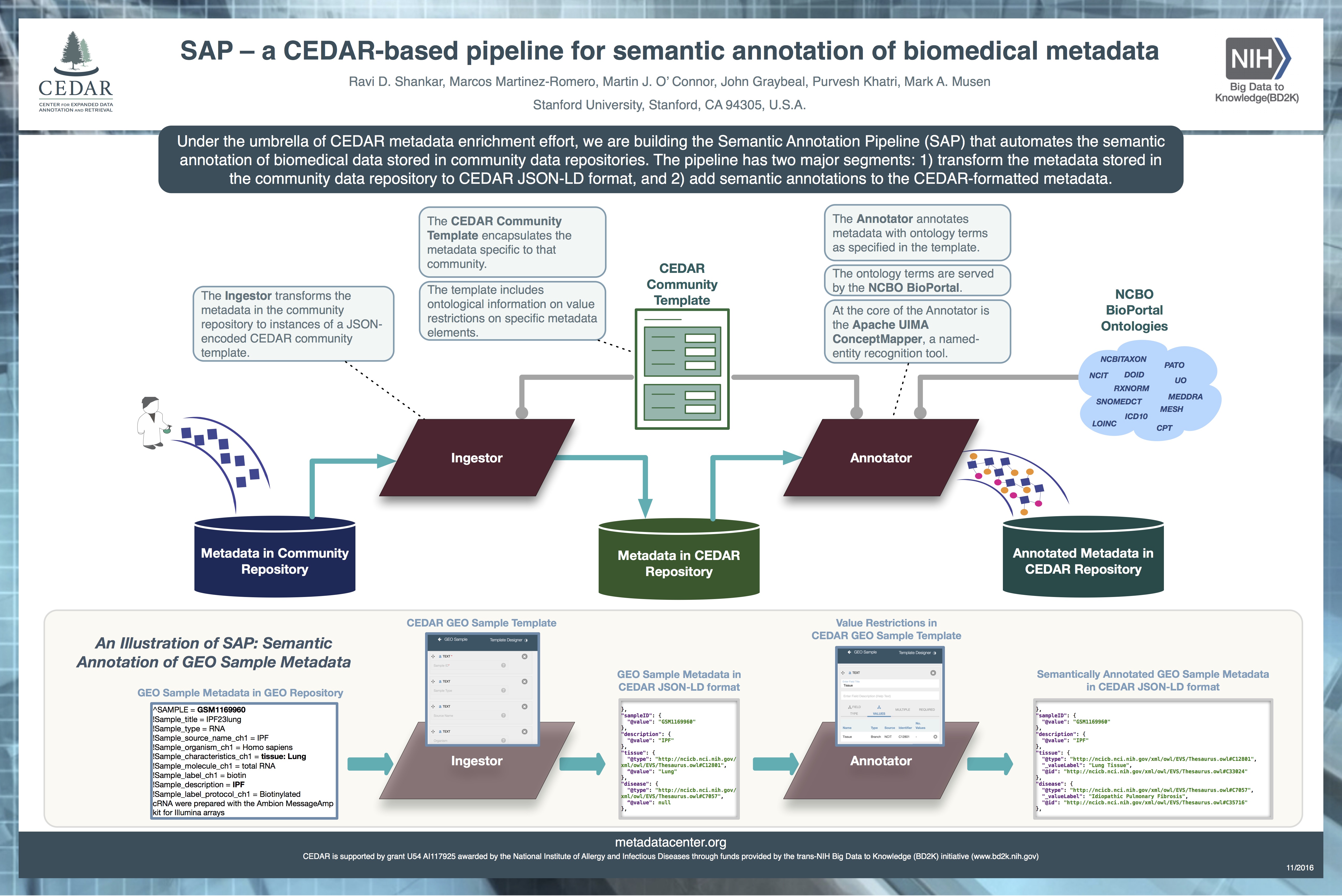
Abstract Title SAP – a CEDAR-based pipeline for semantic annotation of biomedical metadata |
|
|
Abstract Description The exponential growth in the volume of biomedical data held in public data repositories has created tremendous opportunity to evaluate novel research hypotheses in silico. But such search and analysis of disparate data presupposes a consistent semantic representation of the metadata that annotate the research data. Semantic grouping of data is the cornerstone of efficient searches and meta-analyses. Existing metadata are either granularly defined as tag–value pairs (e.g., sample organism="homo sapiens") or implicitly found in long textual descriptions (e.g., in a study design overview). Current practice is to manually map metadata strings to ontological terms before any data analysis can begin. But manual semantic annotation is time-consuming and requires domain and ontology expertise, and therefore may not scale with metadata growth. Under the umbrella of the Center for Enhanced Data Annotation and Retrieval (CEDAR) metadata enrichment effort, we are building the Semantic Annotation Pipeline (SAP), which automates semantic annotation of biomedical data stored in public data repositories. The pipeline has two major segments: 1) Reformat the metadata stored in the data repository to CEDAR JSON-LD format, using templates created in the CEDAR repository, and 2) Add semantic annotations to the CEDAR formatted metadata. We are employing Apache’s UIMA ConceptMapper to efficiently map metadata text segments to ontology terms. We are using the GEO microarray data repository to build and evaluate SAP. Our initial focus is on annotating a specific set of experiment metadata including experiment design, and sample characteristics such as organism, disease, and treatment. We plan to evaluate the SAP annotations against manually curated data, including GEO datasets found in the GEO repository. We intend to show that SAP can ease the process of semantic annotation of metadata, and the enriched metadata can support efficient search and meta-analyses of biological and biomedical data. |
|